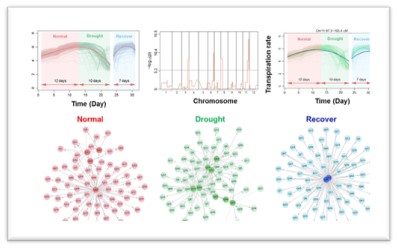
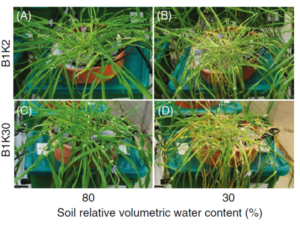
Machine learning (ML) is revolutionizing plant analysis, offering powerful tools to accelerate discovery, optimize resource use, and enhance our understanding of plant behavior. As phenotyping shifts from manual observations to high-resolution, automated systems, ML plays a critical role in extracting meaningful patterns from large, complex datasets.
What Machine Learning Techniques Are Used in Modern Plant Phenotyping?
Machine learning techniques are increasingly applied in plant phenotyping to analyze complex, high-dimensional datasets that describe plant growth, physiology, and environmental interactions. Commonly used approaches include supervised learning methods such as decision trees, Random Forest, XGBoost, and neural networks, as well as unsupervised techniques like principal component analysis and clustering.
Recent research published in Plant, Cell & Environment (2025) demonstrated that tree-based models and gradient boosting methods perform particularly well for predicting whole-plant daily transpiration when trained on long-term physiological datasets (Friedman et al., 2025). Similar approaches have been applied in studies of sap flow, biomass accumulation, and water use efficiency across multiple crop species, showing that machine learning can capture non-linear plant-environment relationships more effectively than traditional empirical models.
| Model | RMSE | R2 | Run time (sec) |
|---|---|---|---|
| Decision tree | 104.51a | 0.83a | 0.01a |
| Random forest | 86.93a | 0.88a | 1.65a |
| XGBoost | 85.19a | 0.89a | 0.24a |
| Neural Network | 84.63a | 0.89a | 57.99a |
Note: Values are averages from 10-fold cross-validation. An ANOVA test was used to evaluate performance differences among models, followed by Tukey’s Honest Significant Difference (HSD). Different letters indicate significant differences. ap < 0.05.
Comparison of machine learning model accuracy and runtime for plant phenotyping based on cross-validation results, highlighting trade-offs between predictive performance and computational efficiency.
Adapted from Friedman et al. Friedman et al. (2025), Integrating Load-Cell Lysimetry and Machine Learning for Prediction of Daily Plant Transpiration.)
How Does Machine Learning Improve Plant Trait Analysis?
Machine learning improves plant trait analysis by enabling the integration of multiple physiological and environmental variables into a single predictive framework. Unlike mechanistic or empirical models that rely on predefined assumptions, ML models learn relationships directly from data, allowing them to adapt to complex and dynamic biological systems.
In this study, machine learning models trained on direct physiological measurements outperformed conventional transpiration estimation methods. Feature importance and SHAP analyses revealed that plant biomass and temperature were the dominant drivers of daily transpiration, providing both predictive accuracy and biological interpretability. Comparable improvements in trait detection have been reported in studies focusing on early stress responses, such as the early detection of Fusarium wilt in tomato plants using water-relation measurements, where physiological signals preceded visible symptoms.
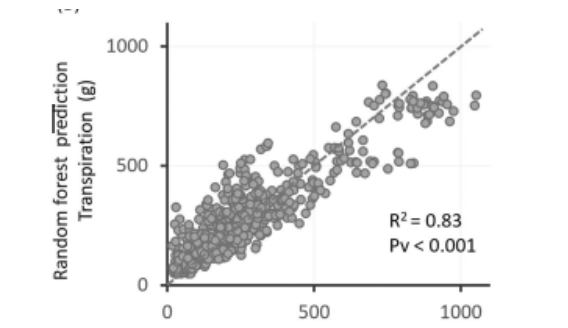
Observed versus Random Forest–predicted daily transpiration showing strong agreement between measured and predicted values (R² = 0.83, p < 0.001); Friedman et al. (2025), Integrating Load-Cell Lysimetry and Machine Learning for Prediction of Daily Plant Transpiration.
What Types of Data Are Most Effective for ML Based Phenotyping?
The effectiveness of machine learning in phenotyping depends strongly on the type and quality of input data. Time-series physiological data, such as transpiration rates, biomass accumulation, and water use efficiency, are particularly valuable because they capture dynamic plant responses over time rather than static snapshots.
Several peer-reviewed studies emphasize the importance of continuous, high-resolution physiological data. For example, Friedman et al. (2025) used seven years of daily physiological and environmental measurements to train robust transpiration prediction models. In another study, Optimized substrate selection for enhanced orchid growth based on high-throughput lysimetric arrays (Yao et al., 2025), continuous physiological measurements enabled quantitative comparison of plant performance across substrates. Together, these studies highlight that machine learning models benefit most from datasets that are continuous, biologically grounded, and collected at the whole-plant level.
How to choose the Right Machine Learning Model for Phenotyping?
Selecting an appropriate machine learning model depends on the research objective, data structure, and desired level of interpretability. Tree-based models such as Random Forest and XGBoost are often used as baseline approaches because they handle non-linear relationships well and provide interpretable feature importance metrics.
In Integrating Load-Cell Lysimetry and Machine Learning for Prediction of Daily Plant Transpiration (2025), these models showed strong generalization when evaluated on independent holdout experiments and external datasets. Neural networks achieved comparable accuracy but required greater computational resources and provided lower transparency. Together, these findings suggest that model selection in plant phenotyping should balance predictive performance with model transparency and biological relevance.
| Model | R2 | RMSE (g) | MAE (g) | Relative error (%) | Residual (g) | Residual (%) | Tukey HSD |
|---|---|---|---|---|---|---|---|
| Decision Tree | 0.77 | 98.65 | 75.53 | 64.71 | 75.53 | 64.71 | a |
What Are the Main Challenges for Phenotyping Plants?
Despite advances in machine learning, several challenges remain in plant phenotyping. Data quality and standardization remain major limitations, particularly when datasets are collected across different environments, seasons, or experimental setups. Sparse measurements, indirect proxies, and sensor noise can reduce the reliability of machine learning predictions.
Another challenge is ensuring that machine learning outputs remain biologically meaningful. Studies such as the Fusarium wilt detection work and the long-term transpiration analysis by Friedman et al. (2025) show that models trained on direct physiological measurements are more likely to produce interpretable and reproducible results. However, generating such datasets at scale remains technically demanding.
Can Machine Learning Support Growth and Yield Prediction?
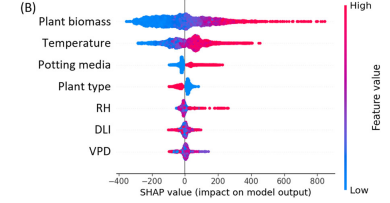
Machine learning has demonstrated potential for supporting growth and yield prediction by linking early physiological traits with later developmental outcomes. While many studies focus on individual traits such as transpiration or biomass, their implications extend to broader performance indicators.
Research using long-term physiological monitoring, including the Friedman et al. (2025), Integrating Load-Cell Lysimetry and Machine Learning for Prediction of Daily Plant Transpiration. study and substrate optimization work by Yao et al. (2025), suggests that early-stage physiological dynamics can inform later growth trajectories. When integrated with environmental data, these approaches may contribute to improved selection strategies in breeding programs and to more informed management decisions.

In this context, platforms such as PlantArray enable the collection of continuous, whole-plant physiological datasets that are suitable for machine learning analysis. Systems developed by Plant-ditech have been used in multiple peer-reviewed studies to support this type of data-driven phenotyping, illustrating how experimental infrastructure can shape the effectiveness of machine learning applications in plant science.